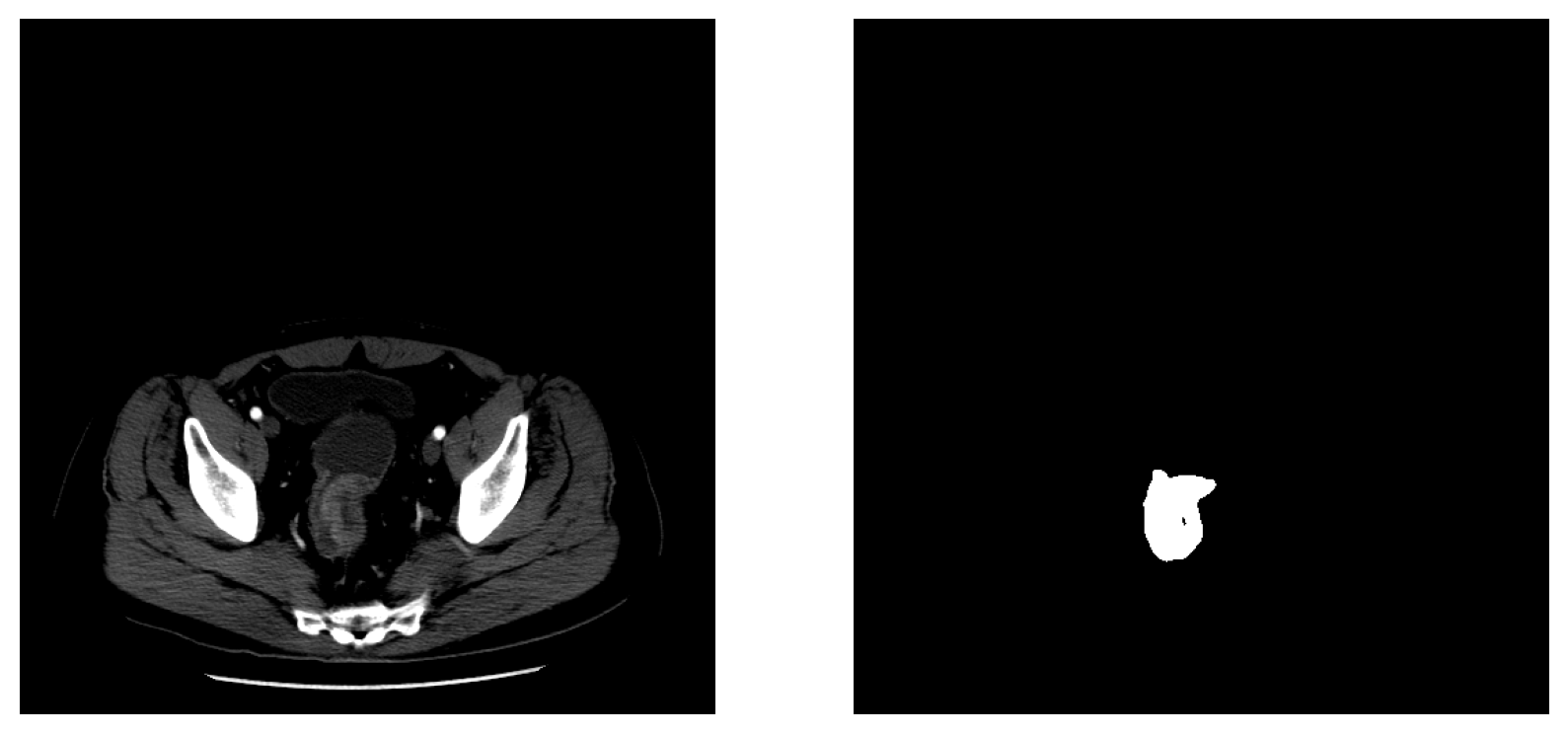
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
|
class Unet(nn.Module):
def __init__(self, in_ch, out_ch):
super(Unet, self).__init__()
self.conv1 = DoubleConv(in_ch, 64)
self.max_pool = nn.MaxPool2d(2)
self.conv2 = DoubleConv(64, 128)
self.conv3 = DoubleConv(128, 256)
self.conv4 = DoubleConv(256, 512)
self.conv5 = DoubleConv(512, 1024)
self.up6 = nn.ConvTranspose2d(1024, 512, 2, stride=2)
self.conv6 = DoubleConv(1024, 512)
self.up7 = nn.ConvTranspose2d(512, 256, 2, stride=2)
self.conv7 = DoubleConv(512, 256)
self.up8 = nn.ConvTranspose2d(256, 128, 2, stride=2)
self.conv8 = DoubleConv(256, 128)
self.up9 = nn.ConvTranspose2d(128, 64, 2, stride=2)
self.conv9 = DoubleConv(128, 64)
self.conv10 = nn.Conv2d(64, out_ch, 1)
def forward(self, x):
c1=self.conv1(x)
p1=self.max_pool(c1)
c2=self.conv2(p1)
p2=self.max_pool(c2)
c3=self.conv3(p2)
p3=self.max_pool(c3)
c4=self.conv4(p3)
p4=self.max_pool(c4)
c5=self.conv5(p4)
up_6= self.up6(c5)
merge6 = torch.cat([up_6, c4], dim=1)
c6=self.conv6(merge6)
up_7=self.up7(c6)
merge7 = torch.cat([up_7, c3], dim=1)
c7=self.conv7(merge7)
up_8=self.up8(c7)
merge8 = torch.cat([up_8, c2], dim=1)
c8=self.conv8(merge8)
up_9=self.up9(c8)
merge9=torch.cat([up_9,c1],dim=1)
c9=self.conv9(merge9)
c10=F.sigmoid(self.conv10(c9))
return c10
|